About BacFITBase
BacFitBase is a manually curated database of bacterial genes that includes information on their relevance during host infection, as measured by transposon mutagenesis. It contains more than 90,000 entries with information on the contribution of individual genes to bacterial fitness under in vivo infection conditions. The data were collected from 15 different studies where transposon mutagenesis was performed. Overall, BacFITBase includes information on 15 pathogenic bacteria and 5 host vertebrates across 10 different tissues.
Why study bacterial fitness in bacterial infection processes?
The development of new antimicrobial therapies relies heavily on our understanding of the mechanisms of bacterial infection. Therefore, it is crucial to understand how bacterial infection develops in vivo and which bacterial genes are required to infect a host. Transposon mutagenesis experiments allow the measurement of fitness values for individual genes, allowing us to assess which genes are fundamental to infect a specific host organism.
Background
To address the contribution of a bacterial gene to infection, its fitness is measured through transposon mutagenesis. Briefly, mutants randomly targeting almost all genes in the bacterial genome are created by insertion of transposable elements, or transposons. Afterwards, these mutants are grown in culture medium (input pool), inoculated to a host organism and finally recovered after infection (output pool). Genomic DNA from the input and output pools is extracted, and transposon insertion site junctions are amplified and quantified by sequencing. Reads for each transposon insertion site are normalized to the total number of reads obtained from that sample (Fig. 1).
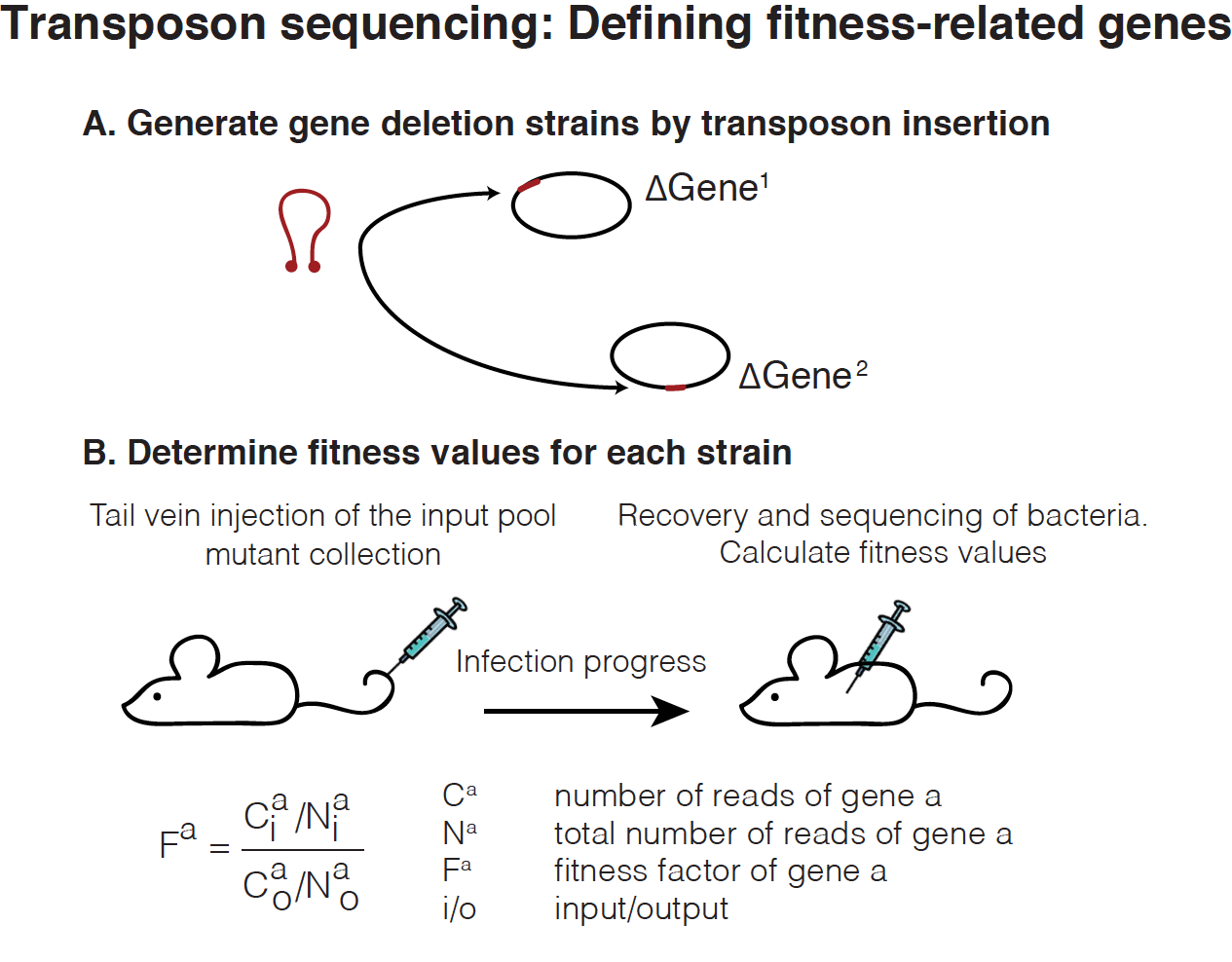
Fitness scores during infection
The fitness score of a gene is calculated as the ratio of normalized frequency of input/output read counts. In this database we collected data from publicly available transposon mutagenesis experiments containing either raw input/output read counts or fitness scores for all mutant genes obtained. In order to standardize all fitness scores, their corresponding z-scores were calculated for each individual experiment (Fig. 2).
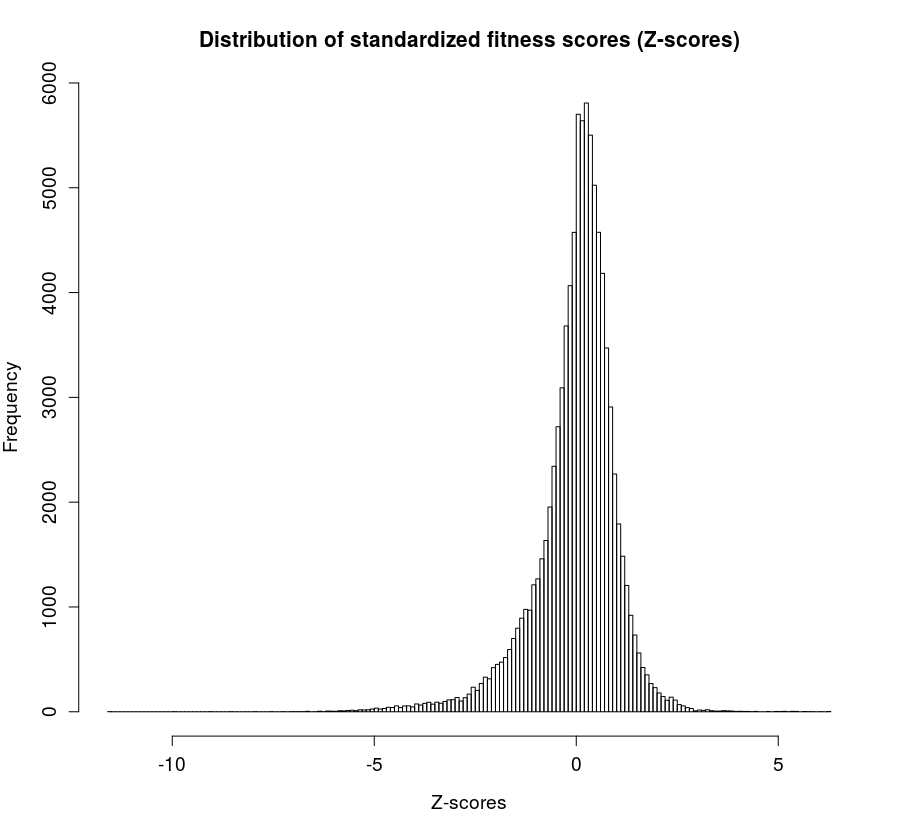
The p-values shown derive from the original publications, where available. For studies that did not provide p-values, we calculated p-values using a two-tailed one-sample Student's t-test on the distribution of fitness scores within each study.
While the z-score and p-values available in our database allow direct comparison between different studies, we recommend referring to the original publications (which are linked to on our data display pages) for a more in-depth analysis.
Updates
You can follow us on Twitter at @tartaglialab and @sysbiogr for updates.Contact
Please feel free to email Javier Macho (javier.macho@uab.cat), Benjamin Lang (benjamin.lang@crg.eu), Gian Gaetano Tartaglia (gian@tartaglialab.com), and Marc Torrent (marc.torrent@uab.cat) — any questions, ideas and feedback are very welcome.How to cite BacFITBase
Please reference Macho Rendón, J., Lang, B., Tartaglia, G.G., and Torrent Burgas, M. (2020). BacFITBase: a database to assess the relevance of bacterial genes during host infection. Nucleic Acids Res. 48, D511–D516.References
- Crua Asensio, N., Muñoz Giner, E., de Groot, N.S., and Torrent Burgas, M. (2017). Centrality in the host-pathogen interactome is associated with pathogen fitness during infection. Nature Communications 8, 14092.
- Jehl, P., Manguy, J., Shields, D.C., Higgins, D.G., and Davey, N.E. (2016). ProViz-a web-based visualization tool to investigate the functional and evolutionary features of protein sequences. Nucleic Acids Res. 44, W11–W15.
Primary data sources
- Anderson, M.T., Mitchell, L.A., Zhao, L., and Mobley, H.L.T. (2017). Capsule Production and Glucose Metabolism Dictate Fitness during Serratia marcescens Bacteremia. MBio 8, 755.
- Bachman, M.A., Breen, P., Deornellas, V., Mu, Q., Zhao, L., Wu, W., Cavalcoli, J.D., and Mobley, H.L.T. (2015). Genome-Wide Identification of Klebsiella pneumoniae Fitness Genes during Lung Infection. MBio 6, e00775.
- Chaudhuri, R.R., Morgan, E., Peters, S.E., Pleasance, S.J., Hudson, D.L., Davies, H.M., Wang, J., van Diemen, P.M., Buckley, A.M., Bowen, A.J., et al. (2013). Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9, e1003456.
- Eckert, S.E., Dziva, F., Chaudhuri, R.R., Langridge, G.C., Turner, D.J., Pickard, D.J., Maskell, D.J., Thomson, N.R., and Stevens, M.P. (2011). Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J Bacteriol 193, 1771–1776.
- Fu, Y., Waldor, M.K., and Mekalanos, J.J. (2013). Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663.
- Gao, B., Vorwerk, H., Huber, C., Lara-Tejero, M., Mohr, J., Goodman, A.L., Eisenreich, W., Galán, J.E., and Hofreuter, D. (2017). Metabolic and fitness determinants for in vitro growth and intestinal colonization of the bacterial pathogen Campylobacter jejuni. PLoS Biol 15, e2001390.
- Gawronski, J.D., Wong, S.M.S., Giannoukos, G., Ward, D.V., and Akerley, B.J. (2009). Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA 106, 16422–16427.
- Grant, A.J., Oshota, O., Chaudhuri, R.R., Mayho, M., Peters, S.E., Clare, S., Maskell, D.J., and Mastroeni, P. (2016). Genes Required for the Fitness of Salmonella enterica Serovar Typhimurium during Infection of Immunodeficient gp91-/- phox Mice. Infect. Immun. 84, 989–997.
- Hubbard, T.P., Chao, M.C., Abel, S., Blondel, C.J., Abel Zur Wiesch, P., Zhou, X., Davis, B.M., and Waldor, M.K. (2016). Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci USA 113, 6283–6288.
- Le Breton, Y., Belew, A.T., Freiberg, J.A., Sundar, G.S., Islam, E., Lieberman, J., Shirtliff, M.E., Tettelin, H., El-Sayed, N.M., and McIver, K.S. (2017). Genome-wide discovery of novel M1T1 group A streptococcal determinants important for fitness and virulence during soft-tissue infection. PLoS Pathog 13, e1006584.
- Miller, D.P., Hutcherson, J.A., Wang, Y., Nowakowska, Z.M., Potempa, J., Yoder-Himes, D.R., Scott, D.A., Whiteley, M., and Lamont, R.J. (2017). Genes Contributing to Porphyromonas gingivalis Fitness in Abscess and Epithelial Cell Colonization Environments. Front Cell Infect Microbiol 7, 378.
- Olson, M.A., Siebach, T.W., Griffitts, J.S., Wilson, E., and Erickson, D.L. (2018). Genome-Wide Identification of Fitness Factors in Mastitis-Associated Escherichia coli. Appl. Environ. Microbiol. 84, 72.
- Subashchandrabose, S., Smith, S.N., Spurbeck, R.R., Kole, M.M., and Mobley, H.L.T. (2013). Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9, e1003788.
- Wang, J., Pritchard, J.R., Kreitmann, L., Montpetit, A., and Behr, M.A. (2014). Disruption of Mycobacterium avium subsp. paratuberculosis-specific genes impairs in vivo fitness. BMC Genomics 15, 415.
- Wang, N., Ozer, E.A., Mandel, M.J., and Hauser, A.R. (2014). Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio 5, e01163–14.
Funding
This study has been funded by the Spanish Ministerio de Ciencia, Innovación y Universidades (SAF2015-72518-EXP, SAF2017-82158-R and RYC-2012-09999) and a Research Grant 2016 by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID).
 This project has also received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 793135.
This project has also received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 793135.
Licence
Our own work is licenced under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International Licence .
.
Acknowledgements
For UniProt proteins, a protein visualisation is automatically generated by ProViz from the Davey lab. ProViz is an interactive exploration tool for investigating the structural, functional and evolutionary features of proteins.NCBI BLAST version 2.9.0+ (March 2019) is used to search by sequence similarity.
Template and CSS from Bootstrap, various small icons from Font Awesome and 'Bacteria' by 'Icons Producer' from the Noun Project, table export to CSV files via ExcellentExport by Jordi Burgos, and table sorting via bootstrap-sortable by Matúš Brliť.
See also: the CRG's legal notice. © 2025 tartaglialab.com